THERYA, 2025, Vol. 16(1):17-27 DOI:10.12933/therya-25-6142 ISSN 2007-3364
Cytogenetic diversity of non-volant small mammals in the Serra dos Órgãos region, Rio de Janeiro state, Brazil
Lena Geise1*, Luciana Guedes Pereira2, Marianne Bello3, and Marcia Aguieiras1
1 Departamento de Zoologia, Universidade do Estado do Rio de Janeiro. Rua São Francisco Xavier, 524. Rio de Janeiro, RJ, Brazil. Email: lenageise@gmail.com; lena.geise@uerj.br (LG) and m.r.aguieiras@gmail.com (MA).
2 KEO International Consultant. Avenida, Dom João II, 11, décimo andar, 1990-221. Lisboa, Portugal. Email: luciana@gpereira.bio.br (LGP).
3 Programa de Pós-Graduação em Ecologia e Evolução, Departamento de Ecologia, Universidade do Estado do Rio de Janeiro. Rua São Francisco Xavier, 524. Rio de Janeiro, RJ, Brazil. E-mail: mariannesbello@gmail.com; bello.marianne@posgraduacao.uerj.br (MB).
* Corresponding author: https://orcid.org/0000-0003-1239-8047.
The Atlantic Forest is one of South America's most biodiverse regions, hosting a significant portion of Brazil's small non-volant mammal diversity, including 267 rodent and 66 marsupial species. The Serra dos Órgãos region in Rio de Janeiro state is a key area for studying this diversity, as it houses 32 rodent and 13 marsupial species. Rodents, unlike marsupials, exhibit a high diversity of chromosomal forms, which serve as important taxonomic tools for identifying cryptic species. Our study used cytogenetic analyses to enhance the taxonomic resolution and understanding of small mammal biodiversity in the Serra dos Órgãos, focusing on the high chromosomal variation in rodents, particularly those within the Sigmodontinae. We collected and karyotyped specimens from 25 localities within the municipalities of Cachoeiras de Macacu, Guapimirim, Petrópolis, and Teresópolis in Rio de Janeiro. These areas include montane and lowland regions of the Serra dos Órgãos, ranging from 100 to 2,100 meters in altitude. Specimens were captured using live traps and handled following ethical guidelines, with karyotypic analysis performed on metaphase chromosomes obtained from bone marrow cell cultures. We analyzed 220 specimens, representing 20 rodent and five marsupial species. Significant intraspecific chromosomal variation was observed in seven rodent species, particularly within the sigmodontines. Akodon cursor displayed variation in fundamental numbers, while Brucepattersonius nebulosus exhibited variation in both diploid and fundamental numbers. New karyotypes were identified for the echimyid Phyllomys spp.. Our findings underscore the importance of cytogenetic analyses in revealing cryptic species and enhancing taxonomic resolution among South American rodents. The chromosomal variation observed highlights the need for integrating cytogenetic data to understand the evolutionary dynamics and biodiversity of the Atlantic Forest.
La Mata Atlántica es una de las regiones más biodiversas de América del Sur, albergando una parte significativa de la diversidad de mamíferos pequeños no voladores de Brasil, incluidos 267 especies de roedores y 66 de marsupiales. La región de Serra dos Órgãos en el estado de Río de Janeiro es un área clave para estudiar esta diversidad, ya que alberga 32 especies de roedores y 13 de marsupiales. Los roedores, a diferencia de los marsupiales, exhiben una alta diversidad de formas cromosómicas, que sirven como herramientas taxonómicas importantes para identificar especies crípticas. Nuestro estudio tiene como objetivo utilizar análisis citogenéticos para mejorar la resolución taxonómica y la comprensión de la biodiversidad de los pequeños mamíferos en la Serra dos Órgãos, centrándose en la alta variación cromosómica en los roedores, particularmente en aquellos dentro de la subfamilia Sigmodontinae. Recogimos y cariotipamos especímenes de 25 localidades dentro de los municipios de Cachoeiras de Macacu, Guapimirim, Petrópolis y Teresópolis en Río de Janeiro. Estas áreas incluyen regiones montañosas y de tierras bajas de la Serra dos Órgãos, que van desde los 100 hasta los 2,100 metros de altitud. Los especímenes fueron capturados utilizando trampas vivas y manejados siguiendo pautas éticas, con análisis cariotípicos realizados en cromosomas metafásicos obtenidos de cultivos de células de médula ósea. Analizamos 220 especímenes, representando 20 especies de roedores y cinco de marsupiales. Se observó una variación cromosómica intraespecífica significativa en siete especies de roedores, particularmente dentro de la familia Cricetidae. Akodon cursor mostró variación en los números fundamentales, mientras que Brucepattersonius nebulosus exhibió variación tanto en los números diploides como en los fundamentales. Se identificaron nuevos cariotipos para Phyllomys spp.. Nuestros hallazgos subrayan la importancia de los análisis citogenéticos para revelar especies crípticas y mejorar la resolución taxonómica entre los roedores sudamericanos. La variación cromosómica observada resalta la necesidad de integrar datos citogenéticos para comprender las dinámicas evolutivas y la biodiversidad de la Mata Atlántica.
Keywords: Atlantic Forest; Didelphimorphia; intraspecific variation; karyotypes; Rodentia.
© 2025 Asociación Mexicana de Mastozoología, www.mastozoologiamexicana.org
Introduction
Among the small non-volant mammals of Brazil, rodents exhibit the greatest diversity, with 267 described species; marsupials are less diverse, with 66 recognized species (Abreu et al. 2023). The Atlantic Forest, considered one of the most diverse regions in South America, harbors a significant portion of this Brazilian diversity (Prado et al. 2015), including 110 rodent and 24 marsupial species. This includes species from open areas at the border of the Atlantic Forest (Brandão and Hingst-Zaher 2021). The Serra dos Órgãos region represents one of the few remaining areas of biodiversity in the state of Rio de Janeiro (Cronemberger and Castro 2009). Serra dos Órgãos National Park houses 32 species of rodents and 13 species of didelphimorph marsupials (Cronemberger et al. 2019).
Unlike didelphimorphs, rodents exhibit a high diversity of chromosomal forms, even within the same genus (e. g., Akodon: Brandão et al. 2021). This variation is evident in diploid and fundamental numbers as well as in the distribution of heterochromatin blocks and other important cytogenetic features (Romanenko and Volobouev 2012). The significant number of karyotype descriptions highlights this high cytogenetic variation, which serves as an important taxonomic tool, especially for genera that contain cryptic species (e. g., Pardiñas et al. 2015; Tribe 2015). Therefore, karyotypic information contributes importantly to the characterization of biodiversity, both at continental scales and within more restricted regions, whether at the individual, population, or higher taxonomic levels (Patterson and Costa 2012). According to Paresque et al. (2018), most karyotypes of Brazilian rodents and marsupials have been available since frequent publication of these data began in the 1970s. However, additional data, particularly when from a considerable number of individuals of the same species from the same locality, offer valuable insights into intraspecific and geographic variation.
The collection efforts of small mammals in the Serra dos Órgãos range began in the 1990s. In 1991, two taxa – Delomys dorsalis and, at the time, a Rhipidomys sp. – were collected in Garrafão (locality 16 of the present work; Figure 1) and karyotyped (L. Geise, personal communication). A long-term study began in 1996, primarily coordinated by the Laboratório de Vertebrados (LabVert, Ecology Department, Universidade Federal do Rio de Janeiro; Gentile and Kajin 2015). This study allowed numerous genetic analyses (Aguieiras et al. 2013; Maestri et al. 2016; Pardiñas et al. 2016; Malcher et al. 2017; Paixão et al. 2021), the description of a new species (Rhipidomys itoan; de Andrade et al. 2011), and a comprehensive species list with their areas of occurrence (Cronemberger et al. 2019). Here, we present a review and broad description of several specimens collected over the past 25 years that have allowed for the acquisition of novel karyotypes.
Materials and methods
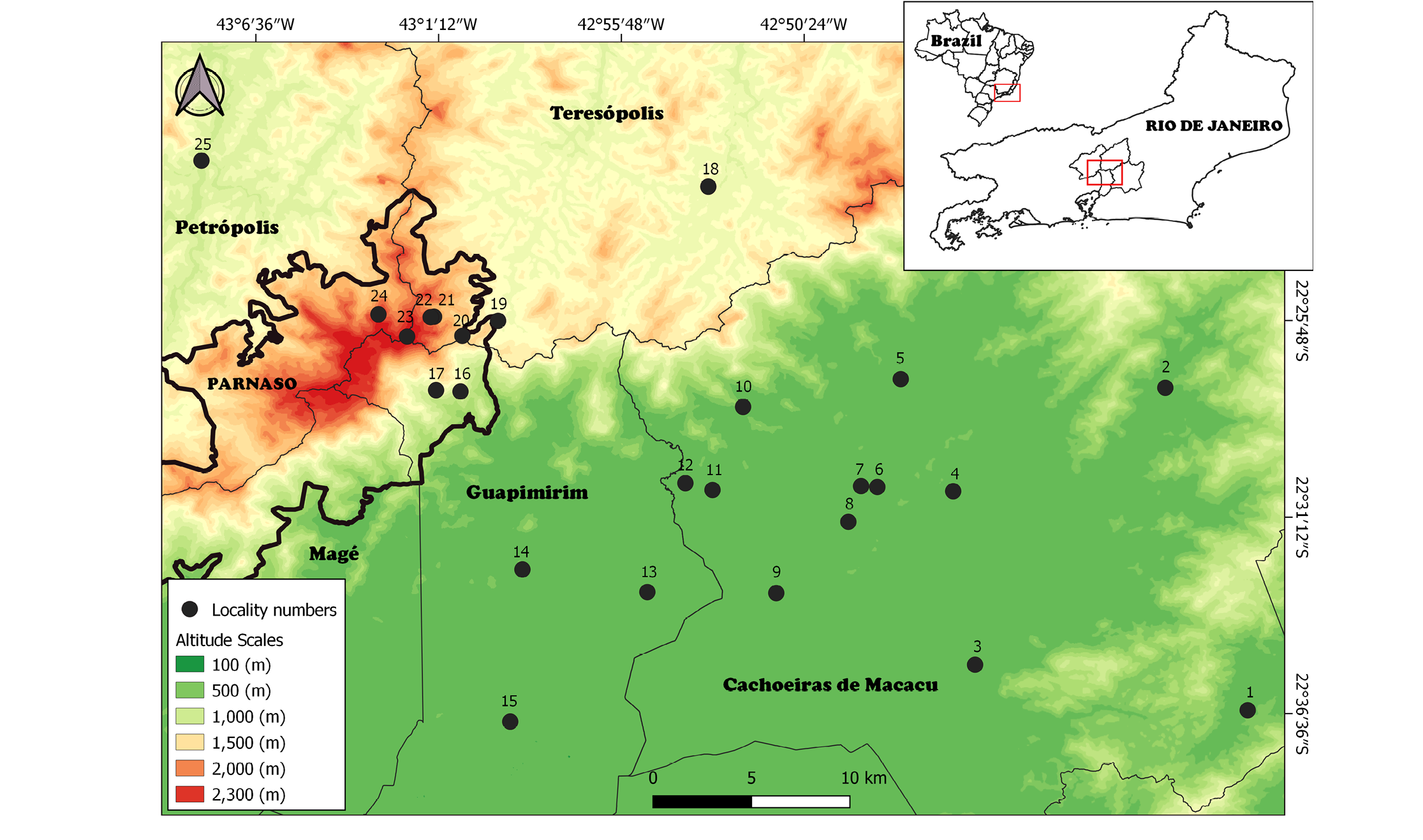
The specimens analyzed for karyotypic data were collected from 25 localities within the municipalities of Guapimirim, Cachoeiras de Macacu, Petrópolis, and Teresópolis, all situated in the state of Rio de Janeiro (Figure 1). These localities are located on the Atlantic slope of the Serra do Mar, encompassing both the hills and lowlands of the Serra dos Órgãos. The mountainous regions (Petrópolis and Teresópolis) are covered by dense submontane and montane rainforest (Rizzini 1954). In contrast, the lowland regions (Guapimirim and Cachoeiras de Macacu) contain fragments of dense ombrophilous forest (tropical rainforest) that are surrounded by pastures or plantations (Cabral and Fiszon 2004). Locations within the Serra dos Órgãos National Park (hereafter PARNASO) range from 400 to 2,100 meters, while those in the fragmented lowland areas are between 100 and 200 meters in altitude. Small terrestrial mammals were collected using live traps (Sherman, Tomahawk, or similar) placed on the ground, in the understory (one to two meters high), or in the canopy (on platforms at least six meters high) from 1991 to 2018.
The small mammals captured were brought to the laboratory and handled according to protocols approved by the American Society of Mammalogists (Sikes et al. 2016). Voucher specimens for all karyotypes were deposited in the mammal collections of Laboratório de Mastozoologia (LabMast, Zoology Department, State University of Rio de Janeiro) and the Museu Nacional (UFRJ). Preliminary identification was based on primary external morphological characteristics. Taxonomic nomenclature follows Astúa (2015) and Faria et al. (2019) for Didelphimorphia, and Patton et al. (2015), Abreu-Júnior and Percequillo (2019), and Abreu et al. (2023) for Rodentia. This taxonomic arrangement differs from https://www.mammaldiversity.org/taxa.html for Guerlinguetus brasiliensis but is in accordance with Abreu et al. (2023).
Metaphase chromosomes were obtained from bone marrow cell cultures following the protocol of Geise (2014), with some preparations including the addition of ethidium bromide. Metaphase preparations were spread on slides, stained with a 5 % Giemsa solution, and examined using a trinocular optical microscope, model Eclipse 50i. For each slide, at least 20 metaphase chromosomes were examined to obtain high-resolution images that allowed for the determination of diploid number (2n), fundamental number (FNa), and chromosomal morphology. Photographs were taken using a Nikon Digital Color DS-Fi1 camera attached to the microscope. Karyotypes were assembled starting with two-armed chromosomes in order of decreasing size, followed by acrocentric chromosomes; each assembly was compared with karyotypes described previously in the literature. The autosomal complement and sex chromosomes were distinguished using the chromosome nomenclature based on centromere position proposed by Levan et al. (1964). Sex chromosomes were identified according to the literature (Table 1). Almost all chromosomal preparations were deposited in the LabMast collection (Geise and Aguieiras 2021).
Results
A total of 220 specimens were karyotyped, comprising 20 rodent species and five marsupials. Among rodents, Cricetidae was the most diverse group sampled, being represented by 15 species from four tribes and one incertae sedis of Sigmodontinae. Other taxa sampled included Echimyidae (three species), Muridae and Sciuridae (one species each; Table 1 and Supplementary Material). Akodon cursor (64 specimens / 15 localities) and A. montensis (24 specimens / 5 localities) were the most intensely sampled species; we karyotyped 73 (from 15 localities) and 34 (from four localities) specimens, respectively. Table 1 provides a detailed summary of all species karyotyped, number of individuals analyzed, observed variation in diploid and fundamental numbers, and form of the sex chromosome pair.
No intraspecific chromosomal variation was detected in the Didelphimorphia sample, which included Caluromys philander (2n = 14, FNa = 20), Gracilinanus microtarsus (2n = 14, FNa = 20), Marmosops incanus (2n = 14, FNa = 24), Monodelphis scalops (2n = 18, FNa = 30), and Philander quica (2n = 22, FNa = 20; Table 1). Similarly, no variation was observed in several rodent species: Akodon montensis (2n = 24, FNa = 42), Castoria angustidens (2n = 46, FNa = 46), Euryoryzomys russatus (2n = 80, FNa = 86), Juliomys ossitenuis (2n = 20, FNa = 36), J. pictipes (2n = 36, FNa = 34), Nectomys squamipes (2n = 56; FNa = 56), Oecomys catherinae (2n = 60, FNa = 62) and Thaptomys nigrita (2n = 52, FNa = 52). We observed a karyotype of 2n = 40, FNa = 74 for the five squirrel specimens (Guerlinguetus brasiliensis) examined. The single specimen of Rattus rattus examined displayed a karyotype of 2n = 38, FNa = 58.
In contrast, intraspecific variation was observed in seven cricetid rodent species; six differed in fundamental number, with the seventh varying in both 2n and FNa (Table 1). All Akodon cursor specimens shared 2n = 14, although their fundamental numbers were 18 (n = 22), 19 (n = 19), 20 (n = 20) and 21 (n = 3; Table 1). Six Brucepattersonius nebulosus specimens (locality 21) displayed 2n = 52, FNa = 54/56 (Figure 2). Oryzomyine species exhibiting intraspecific variation included Oligoryzomys nigripes, which displayed a consistent diploid number (2n = 62) but varied in fundamental number (FNa), with one specimen displaying FNa = 81 and eight specimens displaying FNa = 82. Similarly, Sooretamys angouya showed variation, with two specimens having 2n = 58 and FNa = 60, and one having 2n = 58 and FNa = 61. Other species with karyotypic variation included Rhipidomys itoan, with seven specimens displaying 2n = 44 and FNa = 48, and three specimens displaying 2n = 44 and FNa = 50. For Delomys dorsalis, 12 specimens displayed 2n = 82 and FNa = 80, with four specimens displaying 2n = 82 and FNa = 82 (Table 1). All 14 specimens of Trinomys dimidiatus showed 2n = 60; of the seven specimens of this species that allowed definition of the FNa, six had FNa = 112 and one FNa = 114.
All four specimens of Phyllomys pattoni exhibited 2n = 76. Three of these specimens displayed FNa = 128 (Table 1; Figure 3); this represents the first karyotypic data for this species. Additionally, one specimen (MN84016) identified as Phyllomys sp. by Araújo et al. (2014; = Phyllomys sp. 3) displayed a previously undescribed karyotype of 2n = 52 and FNa = 96 (Figure 4; Table 1).
Discussion
In recent years, the importance of cytogenetic analyses in understanding the biodiversity and evolution of South American small mammals has become increasingly evident. Our study focuses on the non-volant small mammals of the Serra dos Órgãos and surrounding regions, with particular emphasis on karyotypic analysis of specimens from the protected area of PARNASO. Although trapping efforts across the altitudinal range of this area were not equivalent due to varying research goals and logistical constraints, our cytogenetic data have provided crucial insights into the taxonomy and identification of these mammals. This is especially true for the sigmodontine rodents, which present challenges in field identification due to their cryptic nature. Specimens analyzed here were either collected by other researchers who had various non-genetic goals (e. g., Delciellos et al. 2016, 2017, 2019) or were collected according to the geographic characteristics of the region (access, possibility of trapping) and available logistical infrastructure. In general, the purpose of the associated cytogenetic analyses was to assist in the identification of species as part of ecological studies. The collections made in this way reflect the taxonomic challenges noted here - sigmodontine rodents present greater difficulty for identification at the specific level in the field and thus more specimens of this subfamily were collected for identification using karyotypes and other techniques. In the study area, marsupials can be easily identified in the field by their external morphological characteristics. The sampled area has been extensively studied since the 1990s, with long-term monthly collections. As a result, the marsupial fauna is well-known to the point that species can be named based on morphology with a high degree of certainty. Other areas may have marsupials whose species are still under review, which could lead to taxonomic uncertainties. However, the species in Serra dos Órgãos appear to be well established without significant taxonomic doubts— which is why we have had few specimens of this order for cytogenetic studies (Supplementary material).
Another important point that must be highlighted is the interpretation of the morphology of the chromosomes. We consider the fundamental number of Monodelphis scalops to be the same as that in Di-Nizo et al. (2014), which is in accord with Levan et al. (1964) but contrary to Faria et al. (2020), who consider the FNa to be 30. We consider the fundamental number for Gradilinanus microtarsus to be 20, which differs from Pereira et al. (2008), who considered this number to be 24.
According to Abreu et al. (2021), Brucepattersonius nebulosus is the only species in that genus with a FNa differing from that reported by Bonvicino et al. (1998), who found 2n = 52, FNa = 53 in two specimens collected in Itamonte (MG) due to the presence of heteromorphic pairs. Based on the karyotype observed by us for the Serra dos Órgãos sample (2n = 52, FNa = 54/56; locality 21), we confirm that the karyotype of B. nebulosus differs from all other species of the genus (B. griserufescens, B. soricinus and B. iheringi). A different result (2n = 52; FNa = 52) was also found in one specimen collected in the Parque Nacional da Bocaina (Delciellos et al. 2023), which is close to the type locality of B. nebulosus (Abreu-Júnior and Percequillo 2019). Similarly, the variation in the fundamental number described here for Delomys dorsalis (2n = 82 and FNa = 82) adds a new variant to this species, since the only previously known karyotype is 2n = 80 and FNa = 82 (Di-Nizo et al. 2017). We recognize the need for a more detailed chromosomal banding analysis (e. g., Malcher et al. 2017) to gain a clearer and more comprehensive understanding of the chromosomal forms of these taxa and we suggest that data on banding patterns will clarify the chromosomal evolution of these animals (Figure 2).
The high variation in the fundamental number for Trinomys dimidiatus described here is highlighted in Table 1. Banding pattern techniques are also necessary to understand this variation, which may reflect the presence of B chromosomes as suggested by Fagundes et al. (2004) for T. iheringi. Nacif et al. (2023) demonstrated the high taxonomic diversity within this genus, proposing different lineages that probably represent undescribed species, in the southeastern portion of the Brazilian Atlantic Forest.
The genus Phyllomys, one of the most intriguing among echimyid rodents (Araújo et al. 2014), was intensively studied by Leite (2003), who documented the occurrence of more than one species in the Serra dos Órgãos region. Our sample comprises specimens exclusively recorded in the lowest elevations of the area under study (localities 6, 10, and 16, Figure 1), previously identified through DNA analysis (Araújo et al. 2014; Delciellos et al. 2017).
Our four karyotyped specimens of Phyllomys pattoni revealed an undescribed diploid number (2n = 76), with three specimens allowing the determination of a fundamental number of autosomes (FNa) = 128 (Table 1). Emmons et al. (2002) cited a karyotype for P. pattoni (2n = 80, FNa = 112) from a specimen trapped in Espírito Santo state and reported a karyotype of 2n = 72, FNa = 114 for one of the specimens reported here (MN42978, Table 1) via personal communication to one of the authors of this article (L. Geise). However, this information is incorrect, as our extended analysis of this specimen's karyotype could not define the FNa. Moreover, the correct 2n for this specimen is 76, the same found in the three specimens we karyotyped from locality 16, where analysis determined a fundamental number of 128 (Figure 3, Table 1).
Our results serve to correct information provided by Emmons et al. (2002). Our species identifications are robust, having been confirmed by molecular analysis that included samples from various localities and karyotypic forms of this species. It is important to consider that P. pattoni includes individuals with high chromosomal variation, with 2n ranging from 72 to 80 and FNa ranging from 110 to 128, similar to what has been observed in P. nigrispinus (Paresque et al. 2004; Delciellos et al. 2017). Clearly, further studies are needed to clarify the nature and significance of this extensive geographical variation.
Loss and Leite (2011), Araújo et al. (2014), and Abreu-Júnior et al. (2018) identified an undescribed taxon (Phyllomys sp. 3) through molecular analyses. We karyotyped one of the individuals that they examine (MN84016, previously cited by these authors by its field number, FS12-30), revealing a new karyotype for the genus (Table 1). This important finding adds a new diagnostic character for the accurate identification of Phyllomys species, further emphasizing the utility of karyotyping all Atlantic spiny tree rat specimens collected throughout the distribution of the genus. The high morphological (Leite and Patton 2002) and genetic variation observed in this genus needs to be studied in greater detail to better understand the complex evolutionary and biogeographic patterns of South American small mammals.
Cronemberger et al. (2019) provided a list of 45 species of non-volant small mammals (13 marsupials and 32 rodents) recorded in PARNASO. In this study, we present karyotypic data for five marsupials and 20 rodents collected in PARNASO and surrounding areas, covering over half of the species in this protected area. These karyotypic data have facilitated better identification of cryptic species and the description of new taxa. We also emphasize that a correct interpretation of chromosomal morphology is essential for making comparisons between samples, thereby enhancing our understanding of the variation observed among species and across localities.
In conclusion, our study highlights the significant role of cytogenetic analyses in enhancing taxonomic resolution and understanding of biodiversity among South American small mammals. By focusing on the karyotypic diversity within the Serra dos Órgãos region and its surroundings, we have elucidated chromosomal variations and taxonomic complexity within these species. Our findings underscore the importance of integrating different types of data to identify cryptic species and to refine our understanding of the evolutionary and biogeographic dynamics that have shaped the fauna of the Brazilian Atlantic Forest. Continued research utilizing comprehensive cytogenetic and molecular techniques will be essential for uncovering the hidden diversity and evolutionary relationships within this ecologically significant region, ultimately contributing to more informed conservation strategies for the region's unique biodiversity.
Acknowledgments
We want to thank (in alphabetical order) to A. Braz, A. C. Delciellos, B. Torggler, C. E. de V. Grelle, D. Loretto, E. Hingst-Zaher, G. Marroig, H. de G. Bergallo, K. Dinucci, M. Lara, M. Dalloz, M. V. Vieira that made most of the field work. Collecting effort was possible due to the special effort of Rui Cerqueira, Marcus Vinícius Vieira and Carlos Eduardo de Viveiros Grelle. D. Astúa, S. Pavan helped with marsupial identification, and A. C. Loss, J. A. de Oliveira, M. Weksler and Y. L. R. Leite with rodent identification. C. R. Bonvicino, M. Svartman and V. Fagundes with chromosomal morphology interpretation. C. B. Bonvicino and H. N. Seuánez (in memorian) gave institutional support. Authors fellowship includes UERJ (Prociência), CNPq, CAPES (Programa de Pós-graduação em Ecologia e Evolução, PPGEE, UERJ) and Fundação Carlos Chagas Filho de Amparo à Pesquisa do Estado do Rio de Janeiro (FAPERJ) for LG. CNPq, CAPES and UERJ for LGP. CAPES - Finance Code 001 for MSB. CAPES - Finance Code 001 and PROATEC/UERJ for MA.
Literature cited
Abreu. E. F., et al. 2023. Lista de Mamíferos do Brasil (2023-1) [Data set]. Zenodo.
Abreu, E. F., et al. 2021. Systematics of Brucepattersonius Hershkovitz, 1998 (Rodentia, Sigmodontinae): molecular species delimitation and morphological analyses suggest an overestimation in species diversity. Systematics and Biodiversity 19:544–570.
Abreu-Júnior E. F., and A. R. Percequillo. 2019. Small mammals of the Estação Ecológica de Bananal, southeastern Atlantic Forest, Brazil, with description of a new species of Brucepattersonius (Rodentia, Sigmodontinae). Arquivos de Zoologia 50:1–116.
Abreu, M. S. L., et al. 2014. New distribution records of Serra do Mar grass mouse Akodon serrensis Thomas, 1902 (Mammalia: Rodentia: Sigmodontinae) in the southernmost Brazil. Check List 10:655–659.
Abreu-Júnior, E. F., et al. 2018. Unveiling the identity of Kerr’s Atlantic tree rat, Phyllomys kerri (Rodentia, Echimyidae). Mammalian Biology 91:57–70.
Aguieiras. M. R., et al. 2013. Primeiro registro de Juliomys ossitenuis Costa, Pavan, Leite & Fagundes, 2007 e simpatria com Juliomys pictipes (Osgood, 1933) (Rodentia, Cricetidae, Sigmodontinae) na Serra dos Órgãos, Rio de Janeiro. Boletim da Sociedade Brasileira de Mastozoologia 68:57–64.
de Andrade Costa B. M., et al. 2011. Phylogeography of Rhipidomys (Rodentia: Cricetidae: Sigmodontinae) and description of two new species from southeastern Brazil. Journal of Mammalogy 92:945–962.
Araújo N. P., et al. 2014. New karyotypes of Atlantic tree rats, genus Phyllomys (Rodentia: Echimyidae). Genoma 57:1–8.
Astúa, D. 2015. Family Didelphidae (Opossums). Pp. 70–186, in Handbook of the Mammals of the World - Monotremes and Marsupials (Wilson, D. E., and R. A. Mittermeier, org.). Lynx Edicions. Barcelona, España.
Brandão, M. V., and E. Hingst-Zaher. 2021. Atlas Craniano Mamíferos da Mata Atlântica e lista de espécies, 1st ed. TIJD Edições. São Paulo, Brazil
Brandão, M. V., et al. 2021. A new species of Akodon Meyen, 1833 (Rodentia: Cricetidae: Sigmodontinae) endemic from the Brazilian Cerrado. Journal of Mammalogy 102:101–122.
Bonvicino, C. R., V. Penna-Firme, and H. N. Seuánez. 1998. The karyotype of Brucepattersonius griserufescens Hershkovitz, 1998 (Rodentia, Sigmodontiae) with comments on distribution and taxonomy. Zeitschrift für Säugetierkunde 63:329–335.
Bonvicino, C. R., and I. Otazu. 1999. The Wilfredomys pictipes (Rodentia: Sigmodontinae) karyotype with comments on the karyosystematics of Brazilian Thomasomyini. Acta Theriologica 44: 329-332.
Cabral, D. C., and J. T. Fiszon. 2004. Padrões sócio-espaciais de desflorestamento e suas implicações para a fragmentação florestal: Estudo de caso na Bacia do Rio Macacu, RJ. Scientia Forestalis 66:13–24.
Carvalho, B. D. A., et al. 2002. Karyotypes of nineteen marsupial species from Brazil. Journal of Mammalogy 83:58–70.
Christoff, A. U., et al. 2000. Description of a new species of Akodon (Rodentia: Sigmodontinae) from southern Brazil. Journal of Mammalogy 81:838–851.
Costa, B. M. D. A., et al. 2011. Phylogeography of Rhipidomys (Rodentia: Cricetidae: Sigmodontinae) and description of two new species from southeastern Brazil. Journal of Mammalogy 92:945–962.
Cronemberger, C., and E. B. Viveiros de Castro. 2009. The contribution of Serra dos Órgãos National Park to biodiversity conservation. Biodiversity and Land use systems in the fragmented Mata Atlântica of Rio de Janeiro. Göttingen (Civillier Verlag) 93-104.
Cronemberger, C., et al. 2019. Mamíferos do Parque Nacional da Serra dos Órgãos: Atualização da lista de espécies e implicações para a conservação. Oecologia Australis 23:191–214.
Delciellos, A. C., et al. 2017. Syntopy of cryptic Phyllomys (Rodentia: Echimyidae) species: description of the karyotype of Phyllomys nigrispinus and an expansion of the geographic distribution of Phyllomys sulinus. Mammalia 82:266–275.
Delciellos, A. C., et al. 2016. Habitat quality vs. spatial variables as determinants of small mammals’ communities in Atlantic Forest fragments. Journal of Mammalogy 97:253–265.
Delciellos, A. C., et al. 2019. Negative or positive density-dependence in movements depends on climatic seasons: the case of a Neotropical marsupial. Austral Ecology 44:216–222.
Delciellos, A. C., et al. 2023. Updated list of non-volant small mammals from the Serra da Bocaina National Park, southeastern Brazil. Biota Neotropica 23:e20231489-14.
Di-Nizo, C. B., et al. 2014. New karyological data and cytotaxonomic considerations on small mammals from Santa Virginia (Parque Estadual da Serra do Mar, Atlantic Forest, Brazil). Comparative Cytogenetics 8:11–30.
Emmons, L. H., et al. 2002. A review of the named forms of Phyllomys (Rodentia: Echimyidae) with the description of a new species from coastal Brazil. American Museum Novitates 3380:1–40.
Fagundes, V., et al. 1997. ZOO-FISH of a microdissection DNA library and G-banding patterns reveal the homeology between the Brazilian rodents Akodon cursor and A. montensis. Cytogenetic and Genome Research 78:224–228.
Fagundes, V., et al. 2000. X; Y translocation revealed by chromosome microdissection and FISH in fertile XY females in the Brazilian rodent Akodon montensis. Cytogenetic and Genome Research 88:124–129.
Fagundes, V., A. U. Christoff, and Y. Yonenaga-Yassuda. 1998. Extraordinary chromosomal polymorphism with 28 different karyotypes in the neotropical species Akodon cursor (Muridae, Sigmodontinae), one of the smallest diploid numbers in rodents (2n = 16, 15 and 14). Hereditas 129: 263–274.
Fagundes, V., J. P. M. Camacho, and Y. Yonenaga-Yassuda. 2004. Are the dot-like chromosomes in Trinomys iheringi (Rodentia, Echimyidae) B chromosomes? Cytogenetic and Genome Research 106:159–164.
Fagundes, V., et al. 2003. Multiple interstitial ribosomal sites (NORs) in the Brazilian squirrel Sciurus aestuans ingrami (Rodentia, Sciuridae) with 2n = 40: an overview of Sciurus cytogenetics. Genetics and Molecular Biology 26:253–257.
Faria, M. B., R. O. Lanes, and C. R. Bonvicino. 2019. Marsupiais do Brasil. Guia de identificação com base em caracteres morfológicos externos e cranianos, primeira edição. Amélie Editorial. São Caetano do Sul, Brasil.
Faria, M. B., R. O. Lanes, and C. R. Bonvicino. 2 020. Non-volant small mammals (Rodentia and Didelphimorphia) diversity in an isolated area of the Serra da Mantiqueira, Minas Gerais state, Brazil. Boletim do Museu Paraense Emílio Goeldi. Ciências Naturais 15:643–662.
Fernando J. L., and A. Langguth. 2002. Karyotypes of Brazilian squirrels: Sciurus spadiceus and Sciurus alphonsei (Rodentia, Sciuridae). Folia Zoologica 51:201–204.
Geise, L. 2014. Procedimentos genéticos iniciais na captura e preparação de mamíferos. Pp. 194-211, in Técnicas de estudos aplicadas aos mamíferos silvestres brasileiros (Reis, N. R. et al. eds.). 2nd edition, Technical Books Editora. Rio de Janeiro, Brazil.
Geise. L, F. C. Canavez, and H. N. Seuánez. 1998. Comparative karyology in Akodon (Rodentia, Sigmodontinae) from Southeastern Brazil. Journal of Heredity 89:158–163.
Geise, L., and M. Aguieiras. 2021. A coleção de mamíferos do Laboratório de Mastozoologia da Universidade do Estado do Rio de Janeiro. Brazilian Journal of Mammalogy 90:p.e90202104–.
Gentile, R., and M. Kajin. 2015. Estudos empíricos de longo prazo de pequenos mamíferos: a contribuição do professor Rui Cerqueira à biologia de populações. Oecologia Australis 19:1-15.
Gonçalves, P. R., and J. A. de Oliveira. 2014. An integrative appraisal of the diversification in the Atlantic Forest genus Delomys (Rodentia: Cricetidae: Sigmodontinae) with the description of a new species. Zootaxa 3760:1–38.
Leite, Y. L. R. 2003. Evolution and systematics of the Atlantic tree rats, genus Phyllomys (Rodentia, Echimyidae), with description of two new species. University of California publications in zoology 132:1–118.
Leite, Y. L. R., and J. L. Patton. 2002. E volution of South American spiny rats (Rodentia, Echimyidae): the star-phylogeny hypothesis revisited. Molecular Phylogenetics and Evolution 25:455-464.
Levan, A. L., K. Fredga, and A. A. Sandberg. 1964. Nomenclature for centromeric position on chromosomes. Hereditas 52:201–220.
Loss, A. C., and Y. L. R. Leite. 2011. Evolutionary diversification of Phyllomys (Rodentia: Echimyidae) in the Brazilian Atlantic Forest. Journal of Mammalogy 92:1352–1366.
Maestri, R., et al. 2016. Predictors of intraspecific morphological variability in a tropical hotspot: comparing the influence of random and non-random factors. Journal of Biogeography 43:2160–2172.
Malcher, S. M., et al. 2017. Oecomys catherinae (Sigmodontinae, Cricetidae): Evidence for chromosomal speciation? PLoS One 12:p.e0181434.
Nacif, C. L., et al. 2023. Hidden diversity of the genus Trinomys (Rodentia: Echimyidae): phylogenetic and populational structure analyses uncover putative new lineages. Zoological Journal of the Linnean Society 198:113-130.
Oliveira, de J. A., and P. R. Gonçalves. 2015. Genus Oxymycterus. Pp. 247–267, in Mammals of South America. Volume 2: Rodents (Patton, J. L., Pardiñas U. F. J, and D’Elía, G. eds.). University of Chicago Press. Chicago, U.S.A.
Paixão, V. dos S., et al. 2021. Comparative genomic mapping reveals mechanisms of chromosome diversification in Rhipidomys species (Rodentia, Thomasomyini) and syntenic relationship between species of Sigmodontinae. PLoS One 16:p.e0258474–21.
Pardinãs, U. F. J., et al. 2016. A new genus for Habrothrix angustidens, and Akodon serrensis (Rodentia: Cricetidae): again paleontology meets neontology in the legacy of Lund. 2016. Mastozoología Neotropical 23:93–115.
Pardiñas, U. F. J., et al. 2015. Genus Akodon. Pp. 144–203, in Mammals of South America. Volume 2: Rodents (Patton, J. L., U. F. J. Pardiñas, and G. D’Elía, eds.). University of Chicago Press. Chicago, U.S.A.
Paresque, R., et al. 2004. Composição cariotípica da fauna de roedores e marsupiais de duas áreas de Mata Atlântica do Espírito Santo. Brasil. Boletim do Museu de Biologia Mello Leitão 17:5–33.
Paresque, R., et al. 2007. Karyological geographic variation of Oligoryzomys nigripes Olfers, 1818 (Rodentia, Cricetidae) from Brazil. Genetics and Molecular Biology 30:43–53.
Paresque, R., J. da S. Rodrigues, and K. B. Righetti. 2018. Karyotypes of Brazilian non-volant small mammals (Didelphidae and Rodentia): An online tool for accessing the chromosomal diversity. Genetics and Molecular Biology 41:605-610.
Patterson, B., and L. P. Costa (eds.). 2012. Bones, Clones, and Biomes: The History and Geography of Recent Neotropical Mammals, 1st ed. University of Chicago Press. Chicago, U.S.A.
Patton, J. L., U. F. J. Pardiñas, and G. D’Elía (eds.). 2015. Mammals of South America. Volume 2, Rodents, 1st ed. The University of Chicago Press. Chicago, U.S.A.
Pereira, L. G., and L. Geise. 2007. Karyotype composition of some rodents and marsupials from Chapada Diamantina (Bahia, Brasil). Brazilian Journal of Biology 67:509–518.
Pereira, N., et al. 2008. Karyotype characterization and nucleolar organizer regions of marsupial species (Didelphidae) from areas of Cerrado and Atlantic Forest in Brazil. Genetics and Molecular Biology 31:887-892.
Pinheiro, P. S., and L. Geise. 2008. Non-volant mammals of Picinguaba, Ubatuba, state of São Paulo, southeastern Brazil. Boletim Do Museu de Biologia Mello Leitão 23:51–59.
Prado, J. R., et al. 2015. Species richness and areas of endemism of oryzomyine rodents (Cricetidae, Sigmodontinae) in South America: an NDM/VNDM approach. Journal of Biogeography 42:540-551.
Rizzini, C. T. 1954. Flora Organensis. Arquivos do Jardim Botânico do Rio de Janeiro, XVIII, 115– 246.
Romanenko, A. S., and V. Volobouev. 2012. Non-sciuromorph rodent karyotypes in evolution. Cytogenetic and Genome Research 137:233–245.
Sikes, R. S., et al. 2016. Guidelines of the American Society of Mammalogists for the use of wild mammals in research and education. Journal of Mammalogy 97:663–688.
Silva, M. J. de J., and Y. Yonenaga-Yassuda. 1998. Heterogeneity and meiotic behaviour of B and sex chromosomes, banding patterns and localization of (TTAGGG)n sequences by fuorescence in situ hybridization in the neotropical water rat Nectomys (Rodentia, Cricetidae). Chromosome Research 6:455-462
Souza, É. M. S. de., et al. 2013. Variations of chromosomal structures in Caluromys philander (Didelphimorphia: Didelphidae) from the Amazon region. Genetica 141:89–93.
Souza, M. M. de., et al. 2020. New record of Juliomys ossitenuis Costa, Pavan, Leite & Fagundes, 2007 (Rodentia, Sigmodontinae) in Santa Catarina state, southern Brazil. Check List 16:805–809.
Suárez-Villota, E. Y., et al. 2018. Systematics of the genus Oecomys (Sigmodontinae: Oryzomyini): molecular phylogenetic, cytogenetic and morphological approaches reveal cryptic species. Zoological Journal of the Linnean Society 184:182–210.
Tribe, C. J. 2015. Genus Rhipidomys Tschudi, 1845. Pp. 583-617, in Mammals of South America. Volume 2: Rodents (Patton, J. L., U. F. J. Pardiñas, and G. D’Elía, eds.). University of Chicago Press. Chicago, U.S.A.
de Vivo, M., and A. P. Carmignotto. 2015. Family Sciuridae. Pp. 3-47, in Mammals of South America. Volume 2: Rodents (Patton, J. L., U. F. J. Pardiñas, and G. D’Elía, eds.). University of Chicago Press. Chicago, U.S.A.
Associated editors: Marjorie Matocq and Eileen Lacey
Submitted: August 2, 2024; Reviewed: September 14, 2024
Accepted: November 14, 2024; Published on line: January 31, 2025
Supplementary material
List of specimens for each species of Didelphimorphia and Rodentia (in alphabetic order) analyzed in this study. Numbers in parenthesis correspond to localities provided in the legend of Figure 1. MN correspond to Museu Nacional voucher numbers; Field numbers for specimens are: FS and RF - LabVert, HGB-REGUA - H.G. Bergallo, LG – L. Geise, EDH - Erika Hingst-Zaher, DL - Diogo Loretto, and MFD – M.F. Dalloz.
DIDELPHIMORPHIA
Caluromys philander: (20) Male: MN84877
Gracilinanus microtarsus: (20) Female: RF80. Male: RF64
Marmosops incanus: (2) Male: MN79858; (20) Females: MN83650, MN83651. Males: MN83648, MN83649
Monodelphis scalops: (20) Female: MN81902, MN84921. Males: MN81903, MN84965
Philander quica: (20) Female: MN84888
RODENTIA
Family Cricetidae
Akodon cursor: (1) Male: MN83047; (3) Females: MN85056, MN85057; (4) Females: MN85000, MN85028, MN85034, MN85037. Males: MN85029, MN85033; (5) Females: HGB-REGUA1, HGB-REGUA17. Male: HGB-REGUA2; (6) Females: MN85013, MN85015, MN85017. Males: MN85014, MN85016, MN85018, MN85019; (7) Male: FS6-13; (8) Male: FS6-37; (9) Male: FI39; (10) Females: MN85009. Males: MN85007, MN85008, MN85010; (12) Males: FS5-57, FS5-59; (13) Females: MN84991, FS5-36, MN85003, MN85020, MN85023, MN85025. Males: FS5-3, FS5-58, FS5-69, FS8-79, MN85024. (14) Females: MN76474, FS4-20, FS4-31, FS4-33, MN76487, FS4-41, MN76494, MN76497. Males: MN76475, MN76476, MN76479, FS4-06, FS4-25, MN76490, FS4-43, FS4-45, MN76498, MN76499, MN76501, FS4-61, FS4-81; (15) Female: MN85053; (16) Female: MN48055; (17) Male: MN85060.
Akodon montensis: (18) Female: MN31409; (19) Males: MN59110; (20) Females: MN84839, MN84843, MN84852, MN84860, MN84879. Males: MN84838, MN84840, MN84844, MN84847, MN84848, MN84855, MN84858, MN84870, MN84878, MN84881, MN84885, MN84891; (21) Males: MN84451, MN84460, MN84468; (25) Female: EDH63. Male: JDM2.
Brucepattersonius nebulosus: (20) Male: MN84884; (21) Females: MN84452, MN84453, MN84466, MN84467. Males: MN84464, MN84465.
Castoria angustidens: (21) Females: MN84454, MN84463, MN84469. Males: MN84455, MN84456, MN84461, MN84462; (24) Females: MN69807, MN69812, MN77117. Males: MN77099; Indeterminate: MN77078, MN77079, MN77084, MN77097
Delomys dorsalis: (20) Females: MN84836, MN84841, MN84845, MN84851, MN84859, MN84846, MN84862, MN84865. Males: MN84837, MN84850, MN84853, MN84857, MN84861, MN84863, MN84866; (21) Male: MN84459.
Euryoryzomys russatus: (15) Females: MN71798, MN48023. Male: MN71797.
Juliomys ossitenuis: (19) Female: MN81077. Male: MN81078; (20) Females: MN81083, MN81084, MN81088, MN81092. Males: MN81079, MN81080, MN81081, MN81082, MN81085, MN81086, MN81087, MN81089, MN81090, MN81091; (21) Male: MN84458.
Juliomys pictipes: (11) Male: MFD2; (16) Males: MN81095, MN81096; (20) Female: MN81097; (22) Male: MN81094.
Nectomys squamipes: (10) Males: MN67049.
Oecomys catherinae: (10) Males: MN74373; (11) Males: MFD1.
Oligoryzomys nigripes: (1) Male: MN83048; (5) Female: HGB-REGUA14; (19) Female: MN67472; (20) Female: MN81917. Males: MN84892, MN81916, MN81921, MN81938, MN84912, MN84913, RF1411; (17) Male: MN69888.
Oxymycterus sp: (23) Male: MN48063.
Rhipidomys itoan: (15) Female: HGB 398; (16) Females: MN46805, MN63626, MN63016. Males: MN46801, MN63605; (20) Females: MN81934, MN81935. Males: MN81932, MN84923.
Sooretamys angouya: (20) Females: MN84966, MN84970.
Thaptomys nigrita: (19) Female: MN69838; (20) Males: MN84883, MN84893.
Family Echimyidae
Phyllomys pattoni: (10) Female: MN42978; (16); Females: DL19, DL20. Male: DL21.
Phyllomys sp.: (6) Males: MN84016.
Trinomys dimidiatus: (10) Male: MN67511; (19) Males: MN67503, MN67504; (20) Females: MN84842, MN84849, MN84854, MN84867, MN84871. Males: MN84869, MN84872, MN84873, MN84876, MN84875, MN81087.
Family Muridae
Rattus rattus: (20) Male: MN84962.
Family Sciuridae
Guerlinguetus brasiliensis: (19) Male: MN69839; (16) Female: MN69865; (20) Females: MN81906, MN81907. Male: MN81904.

Figure 1. Collection localities for all karyotyped small terrestrial mammals in the Serra do Mar, Rio de Janeiro state. Lines indicate municipality limits. Colors represent the altitudinal scales, from green (lower altitudes) to red (higher altitudes). Red boxes in both Brazil and Rio de Janeiro State delineate the study area. Locality numbers refer to: Cachoeiras de Macacu Municipality: 1. Fazenda Nova Miracema (-22° 36' 33.7" S, -42º 36' 11.6" W, 102 m); 2. Fragmento (-22° 27′ 50″ S, -42° 39′ 10″ W, 68 m); 3. Fazenda Santo Estevão e Propriedade do Sr. Edimar (-22° 35’ 44” S, -42° 44’ 20” W, 100 m); 4. Fazenda Pica Pau Amarelo (-22° 31’ 00” S, -42º45’ 16” W, 200 m); 5. Reserva Ecológica de Guapiaçu (-22° 28’ 00” S, -42° 46’ 60” W, 34 m); 6. Conjunto de Fazendas (-22° 31’ 00” S, -42° 47’ 31” W, 100 m); 7. Fazenda Sem Nome (-22° 31’ 00” S, -42º48’ 00” W, 150m); 8. Fazenda Parahy (-22° 32’ 00” S, -42º48’ 19” W, 150 m); 9. São José da Boa Morte (-22° 34’ 4” S, -42° 50’ 20” W, 100 m); 10. Sítio Rosimery (-22° 29’ 00” S, -42º51’ 37” W, 200 m); 11. Estação Ecológica do Paraíso (-22º31’ 19.80” S, -42º52’ 23.20” W, 68 m). Guapimirim Municipality: 12. Fazenda Iguaçu (-22° 31’ 11” S, -42º53’ 12” W, 100 m); 13. Fazendas Consorciadas (-22° 34’ 14” S, -42° 54’ 9” W, 150 m); 14. Fazenda Chorona (-22° 33’ 48” S, -42° 57’ 53” W, 200 m); 15. Centro de Primatologia do Rio de Janeiro (INEA) (-22° 38’ 00” S, -42° 58’ 00” W, 100 m); 16. Parque Nacional da Serra dos Órgãos (PARNASO), Garrafão (-22° 29’ 00” S, -43° 00’ 00” W, 700 m); 17. PARNASO, Subsede, Município de Guapimirim (-22° 29’ 00” S, -43° 00’ 43.48” W, 400 m). Teresópolis Municipality: 18. Fazenda Boa Fé (-22° 23’ 00” S, -42° 53’ 00” W, 880 m); 19. PARNASO, Sede (-22° 27’ 00” S, -42° 59’ 00” W, 1,200 m); 20. PARNASO, Rancho Frio (-22° 27’ 28” S, -43º00’ 2” W, 1,200 m); 21. PARNASO, Trilha do Sino, Parte Baixa (-22° 26’ 59” S, -43° 00’ 54” W, 1,627m); 22. PARNASO, Abrigo 4 (-22° 27’ 00” S, -43º01’ 00” W, 2,130 m); 23. PARNASO, Pedra do Sino (-22° 27’ 34” S, -43° 01’ 40” W, 2,100 m). Petrópolis Municipality: 24. PARNASO, Vale das Antas, Município de (-22° 27’ 00” S, -43° 02’ 33” W, 1950 m); 25. Fazenda Inglesa (-22° 23’ 00” S, -43° 08’ 00” W, 910 m).
Table 1. List of species for which karyotypic data were obatined, including diploid chromosome number (2n) and fundamental autosomal number (FNa), sex pair (X and Y), locality number, and reference publications used for karyotype characterization. Legends: Locality numbers are described in the legend of Figure 1. Chromosome morphology: d = dot chromosome, la = large acrocentric, lm = large metacentric, lsm = large submetacentric, ma = medium acrocentric, mm = medium metacentric, msm = medium submetacentric, sa = short acrocentric, sm = short metacentric. Items in bold indicate new results or results that differ from the literature. An * in the references indicates results that differ from the present study.
|
TAXON |
n |
2n |
FNa |
X |
Y |
(Locality) Specimen number |
Karyotype references |
|
ORDER DIDELPHIMORPHIA |
|||||||
|
Caluromys philander |
1 |
14 |
20 |
sa |
d |
(20) MN84877 |
Souza et al. 2013 |
|
Gracilinanus microtarsus |
2 |
14 |
20 |
sm |
d |
(20) RF64, RF80 |
Pereira and Geise 2007 |
|
Marmosops incanus |
5 |
14 |
24 |
mm |
sa |
(2) MN79858; (20) MN83648, MN83649, MN83650, MN83651 |
Carvalho et al. 2002, Paresque et al. 2004, Di-Nizo et al. 2014 |
|
Monodelphis scalops |
4 |
18 |
30 |
sa |
d |
(20) MN81902, MN81903, MN84921, MN84965 |
Di Nizo et al. 2014 |
|
Philander quica |
1 |
22 |
20 |
sa |
- |
(20) MN84888 |
Carvalho et al. 2002, Faria et al. 2020 |
|
ORDER RODENTIA |
|||||||
|
Family Cricetidae |
|||||||
|
Tribe Akodontini |
|||||||
|
Akodon cursor |
22 |
14 |
18 |
sa |
d |
(4) MN85028, MN85034; (5) HGB-REGUA1, HGB-REGUA2, HGB REGUA17; (6) MN85013; (9) FI39; (13) MN85023, MN85025, MN84991, FS5-36, FS5-58, FS5-69, FS8-79; (14) MN76479, MN76487, MN76490, MN76497, MN76498, FS4-61; (16) MN48055; (17) MN85060 |
Fagundes et al. 1998, Geise et al. 1998, Faria et al. 2020 |
|
19 |
14 |
19 |
sa |
d |
(1) MN83047; (3) MN85056, MN85057; (6) MN85016, MN85018, MN85019; (10) MN85007, MN85008; (12) FS5-59; (13) MN85024, FS5-3; (14) MN76494, MN76499, MN76474, MN76475, FS4-20, FS4-41, FS4-43, FS4-45 |
Fagundes et al. 1998, Geise et al. 1998, Faria et al. 2020 |
|
|
19 |
14 |
20 |
sa |
d |
(4) MN85000, MN85029, MN85033, MN85037; (6) MN85014, MN85015; (7) FS6-13; (8) FS6-37; (10) MN85009, MN85010; (12) FS5-57; (13) MN85003; (14) MN76501, MN76476, FS4-6, FS4-25, FS4-31, FS4-33; (15) MN85053 |
Fagundes et al. 1998, Geise et al. 1998, Faria et al. 2020 |
|
|
3 |
14 |
21 |
sa |
d |
(6) MN85017; (13) MN85020; (14) FS4-81 |
Fagundes et al. 1998, Geise et al. 1998, Faria et al. 2020 |
|
|
Akodon montensis |
23 |
24 |
42 |
ma |
sa |
(18) MN31409; (19) MN59110; (20) MN84838, MN84839, MN84840, MN84843, MN84844, MN84847, MN84848, MN84852, MN84855, MN84858, MN84860, MN84870, MN84878, MN84879, MN84881, MN84885, MN84891; (21) MN84451, MN84460, MN84468; (25) EDH63 |
Fagundes et al. 1997*, Geise et al. 1998*, Fagundes et al. 2000 |
|
1 |
24 |
42 |
ma |
sm |
(25) JDM2 |
Fagundes et al. 1997*, Geise et al. 1998*, Fagundes et al. 2000 |
|
|
Brucepattersonius nebulosus |
4 |
52 |
54 |
la |
sa |
(21) MN84453, MN84452, MN84466, MN84467 |
Present study |
|
2 |
52 |
56 |
msm |
sm |
(21) MN84464, MN84465 |
Present study |
|
|
1 |
52 |
- |
- |
- |
(20) MN84884 |
- |
|
|
Castoria angustidens |
12 |
46 |
46 |
ma |
sa |
(21) MN84454, MN84455, MN84456, MN84462, MN84463, MN84461, MN84469; (24) MN69812; (24) MN77078, MN77079, MN77084, MN77097 |
Christoff et al. 2000*, Abreu et al. 2014*, Pardiñas et al. 2015*, 2016* |
|
3 |
46 |
- |
- |
- |
(24) MN69807, MN77099, MN77117 |
- |
|
|
Thaptomys nigrita |
3 |
52 |
52 |
ma |
sm |
(19) MN69838; (20) MN84883, MN84893 |
Yonenaga 1975, Faria et al. 2020* |
|
Oxymycterus sp. |
1 |
54 |
62 |
la |
sa |
(23) MN48063 |
Svartman and Cardoso de Almeida 1993, Oliveira and Gonçalves 2015* |
|
Tribe Oryzomyini |
|||||||
|
Euryoryzomys russatus |
2 |
80 |
86 |
lsm |
sa |
(15) MN71797, MN71798 |
Di-Nizo et al. 2014 |
|
1 |
80 |
- |
- |
- |
(15) MN48023 |
- |
|
|
Nectomys squamipes |
1 |
56 |
56 |
lsm |
sa |
(10) MN67049 |
Silva and Yonenaga-Yassuda 1998 |
|
Oecomys catherinae |
1 |
60 |
62 |
lsm |
la |
(11) MFD1 |
Malcher et al. 2017 |
|
1 |
60 |
- |
- |
- |
(10) MN74373 |
- |
|
|
Oligoryzomys nigripes |
1 |
62 |
81 |
lsm |
sa |
(20) RF1411 |
Faria et al. 2020 |
|
6 |
62 |
82 |
lsm |
ma |
(1) MN83048; (20) MN81916, MN81917, MN81938, MN84892, MN84912 |
Faria et al. 2020 |
|
|
2 |
62 |
82 |
lm |
ma |
(20) MN81921, MN84913 |
Faria et al. 2020 |
|
|
3 |
62 |
- |
- |
- |
(5) HGB-REGUA14; (19) MN67472; (17) MN69888 |
- |
|
|
Sooretamys angouya |
2 |
58 |
60 |
la |
- |
(20) MN84966 |
Di-Nizo et al. 2014, Faria et al. 2020 |
|
1 |
58 |
61 |
la |
- |
(20) MN84970 |
Di-Nizo et al. 2014, Faria et al. 2020 |
|
|
Tribe Thomasomyini |
|||||||
|
Rhipidomys itoan |
7 |
44 |
48 |
lsm |
sa |
(16) MN46801, MN46805, MN63626, MN63016, MN63605; (20) MN81934, MN81935 |
Pinheiro and Geise 2008*, Costa et al. 2011 |
|
3 |
44 |
50 |
lsm |
ma |
(15) HGB398; (20) MN81932, MN84923 |
Pinheiro and Geise 2008*, Costa et al. 2011 |
|
|
Tribe Wiedomyini |
|||||||
|
Juliomys ossitenuis |
1 |
20 |
36 |
lm |
sa |
(19) MN81077, MN81078; (20) MN81079, MN81080, MN81081, MN81082, MN81083, MN81084, MN81085, MN81086, MN81087, MN81088, MN81089, MN81090, MN81091, MN81092; (21) MN84458 |
Aguieiras et al. 2013, Souza et al. 2020 |
|
Juliomys pictipes |
4 |
36 |
34 |
ma |
sa |
(11) MFD2; (16) MN81095, MN81096; (20) MN81097 |
Bonvicino and Otazu 1999, Di-Nizo et al. 2014 |
|
1 |
36 |
- |
- |
- |
(22) MN81094 |
- |
|
|
Incertae Sedis |
|||||||
|
Delomys dorsalis |
13 |
82 |
80 |
lm |
sa |
(20) MN84837, MN84841, MN84845, MN84850, MN84851, MN84853, MN84859, MN84861, MN84863, MN84864, MN84865, MN84866; (21) MN84459 |
Gonçalves and Oliveira 2014* |
|
3 |
82 |
82 |
lm |
sa |
(20) MN84836, MN84857, MN84862 |
Present study |
|
|
Family Echimyidae |
|||||||
|
Trinomys dimidiatus |
6 |
60 |
112 |
lsm |
sa |
(19) MN67503; (20) MN81087, MN84842, MN84849, MN84854, MN84867 |
Present study |
|
1 |
60 |
114 |
lsm |
sa |
(10) MN67511 |
Delciellos et al. 2023 |
|
|
7 |
60 |
- |
- |
- |
(19) MN67504; (20) MN84869, MN84871, MN84872, MN84873, MN84874, MN84875 |
- |
|
|
Tribe Echimyini |
|||||||
|
Phyllomys pattoni |
3 |
76 |
128 |
lm |
sa |
(16) DL19, DL20, DL21 |
Present study |
|
1 |
76 |
- |
- |
- |
(10) MN42978 |
Present study |
|
|
Phyllomys sp. |
1 |
52 |
96 |
lm |
d |
(6) MN84016 |
Present study |
|
Family Muridae |
|||||||
|
Rattus rattus |
1 |
38 |
58 |
la |
sa |
(20) MN84962 |
Kasahara and Yonenaga-Yassuda 1981 |
|
Family Sciuridae |
|||||||
|
Guerlinguetus brasiliensis |
4 |
40 |
74 |
lsm |
ma |
(19) MN69839; (16) MN69865; (20) MN81904, MN81907 |
Fagundes et al. 2003 |
|
1 |
40 |
- |
- |
- |
(20) MN81906 |
- |
|
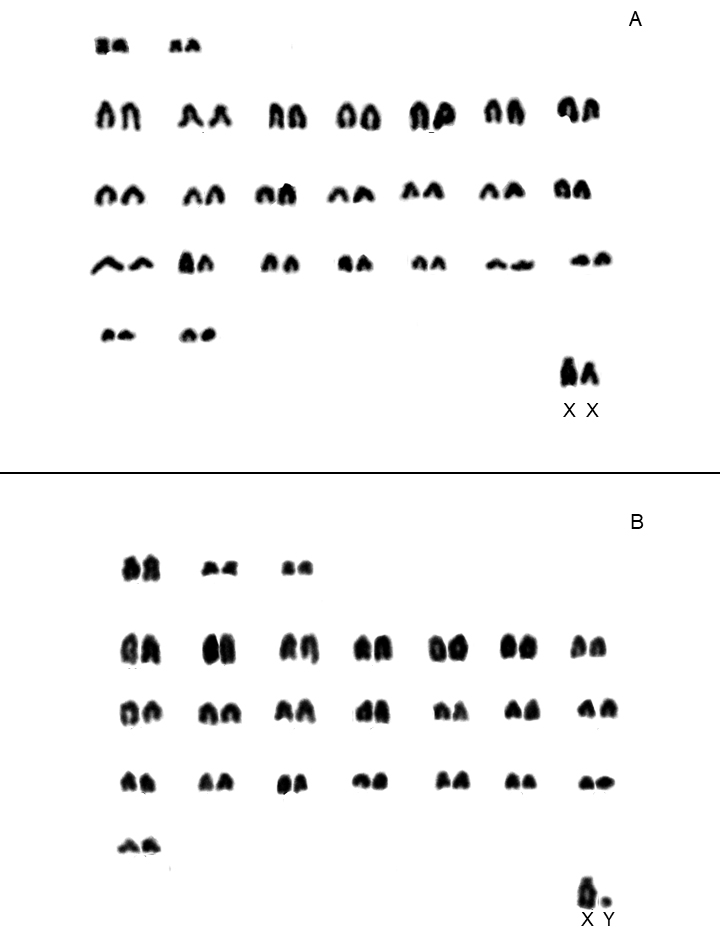
Figure 2. Karyotype of Brucepattersonius nebulosus specimens (A = MN84466) and B = MN84465) obtained using conventional staining. 2n = 52, FNa = 54 and 2n = 52, FNa = 56, respectively.
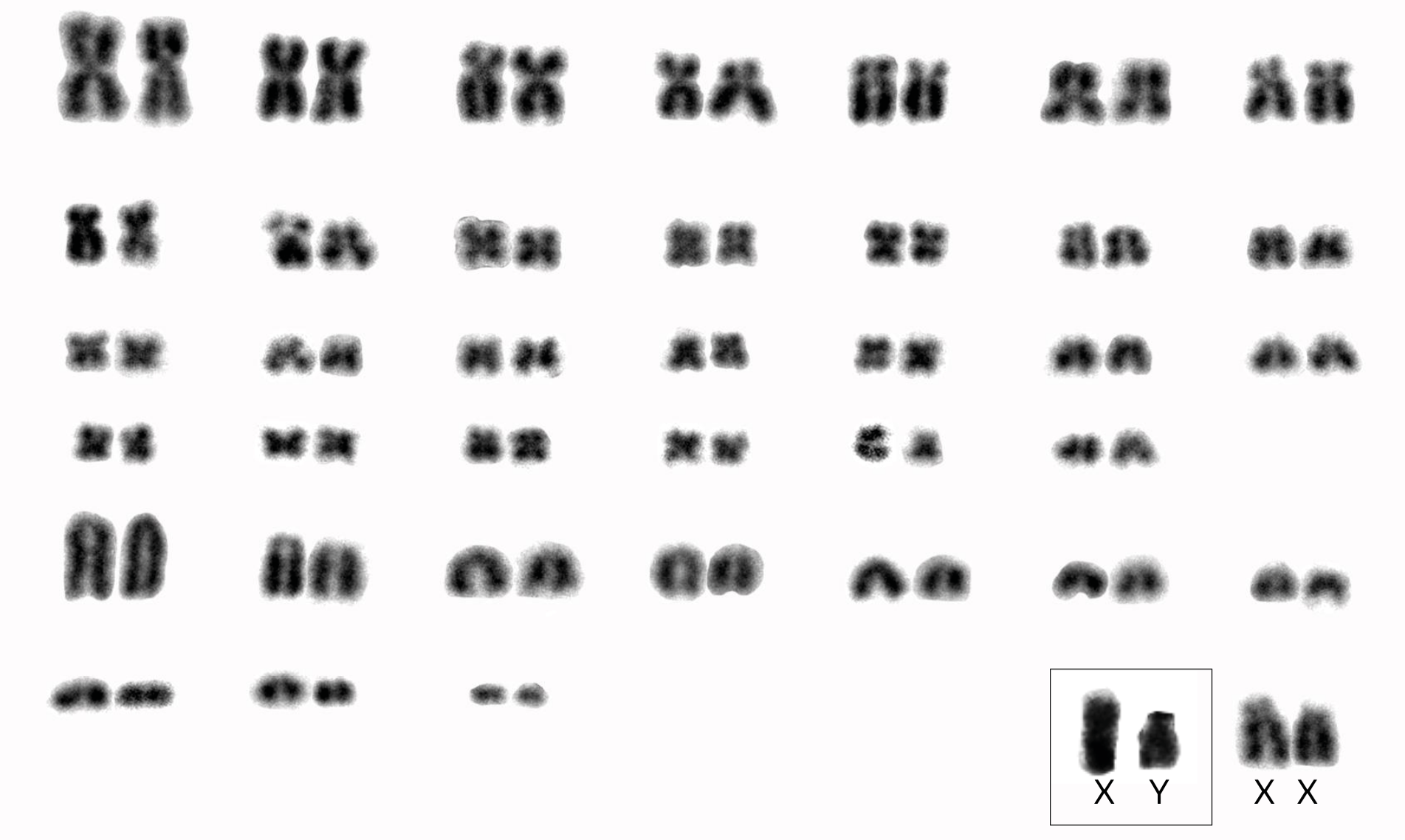
Figure 3. Karyotype a of a Phyllomys pattoni specimen (DL19, female) obtained using conventional staining, 2n = 76, FNa = 128. Sex chromosomes from a male are from DL21.
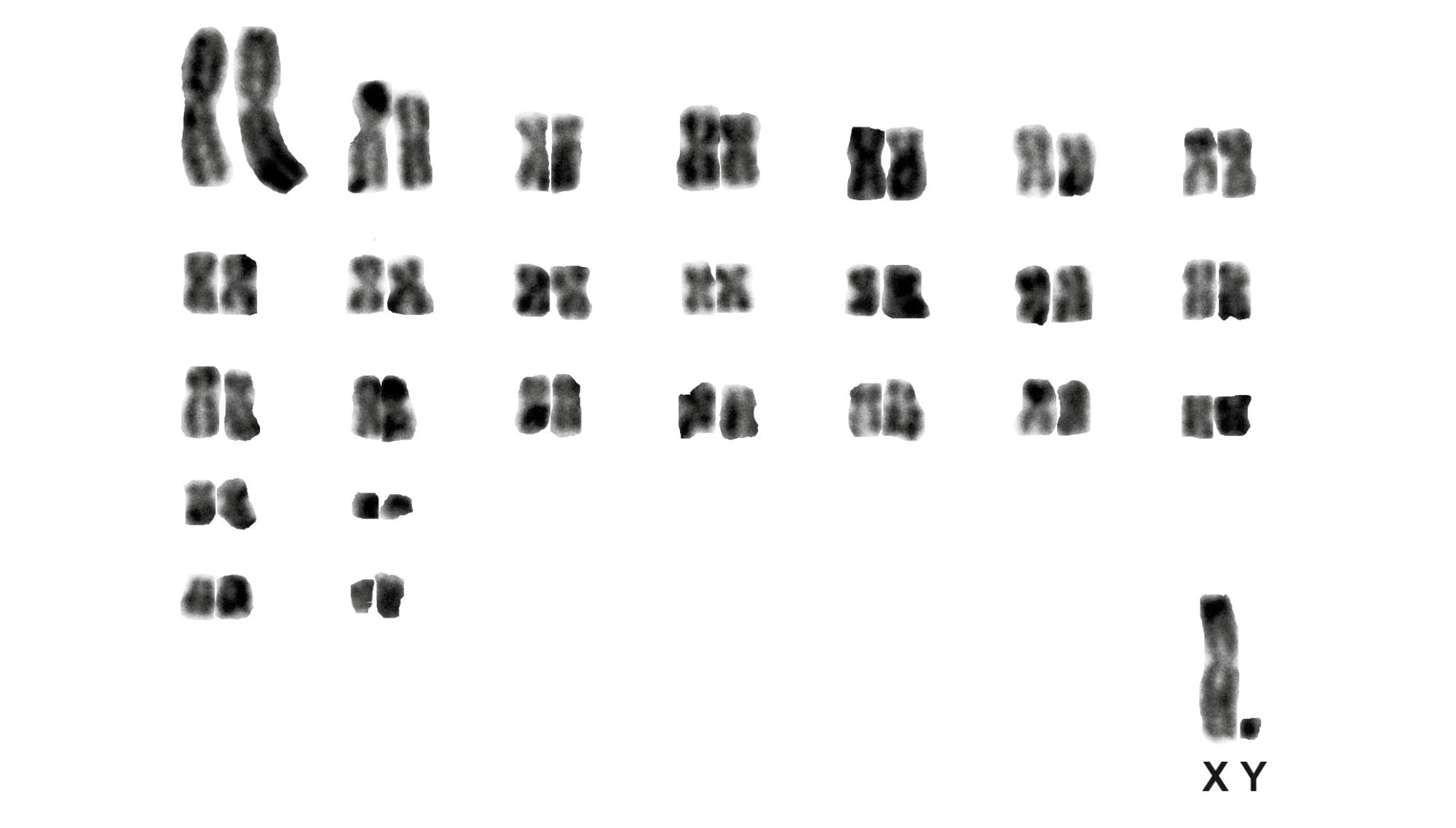
Figure 4. Karyotype of Phyllomys sp. (MN84016) obtained using conventional staining, 2n = 52, FNa = 96.